bigmap-solid-state-nmr
Solid state NMR visualization
Short summary
The goal of this project is to provide an open source microservice (app) for the processing, visualization and analysis of solid-state NMR spectra accessible via a modern web front-end (React).
The project is open-source (MIT license) and is available on https://git.nmrium.org. An exhaustive documentation can be found on https://docs.nmrium.org.
In order to show the possibilities, we provide here a solid state NMR dataset coming from the following publication:
Halat, D., Dunstan, M., Gaultois, M., Britto, S., & Grey, C. (2018). Research data supporting “Study of defect chemistry in the system La 2- Sr NiO 4+ by 17 O solid-state NMR spectroscopy and Ni K-edge XANES” https://doi.org/10.17863/CAM.26025.
It can be previewed from this link https://www.nmrium.org/nmrium#?toc=https%3A%2F%2Fnmrdata.github.io%2Fbigmap-solid-state-nmr%2Ftoc.json.
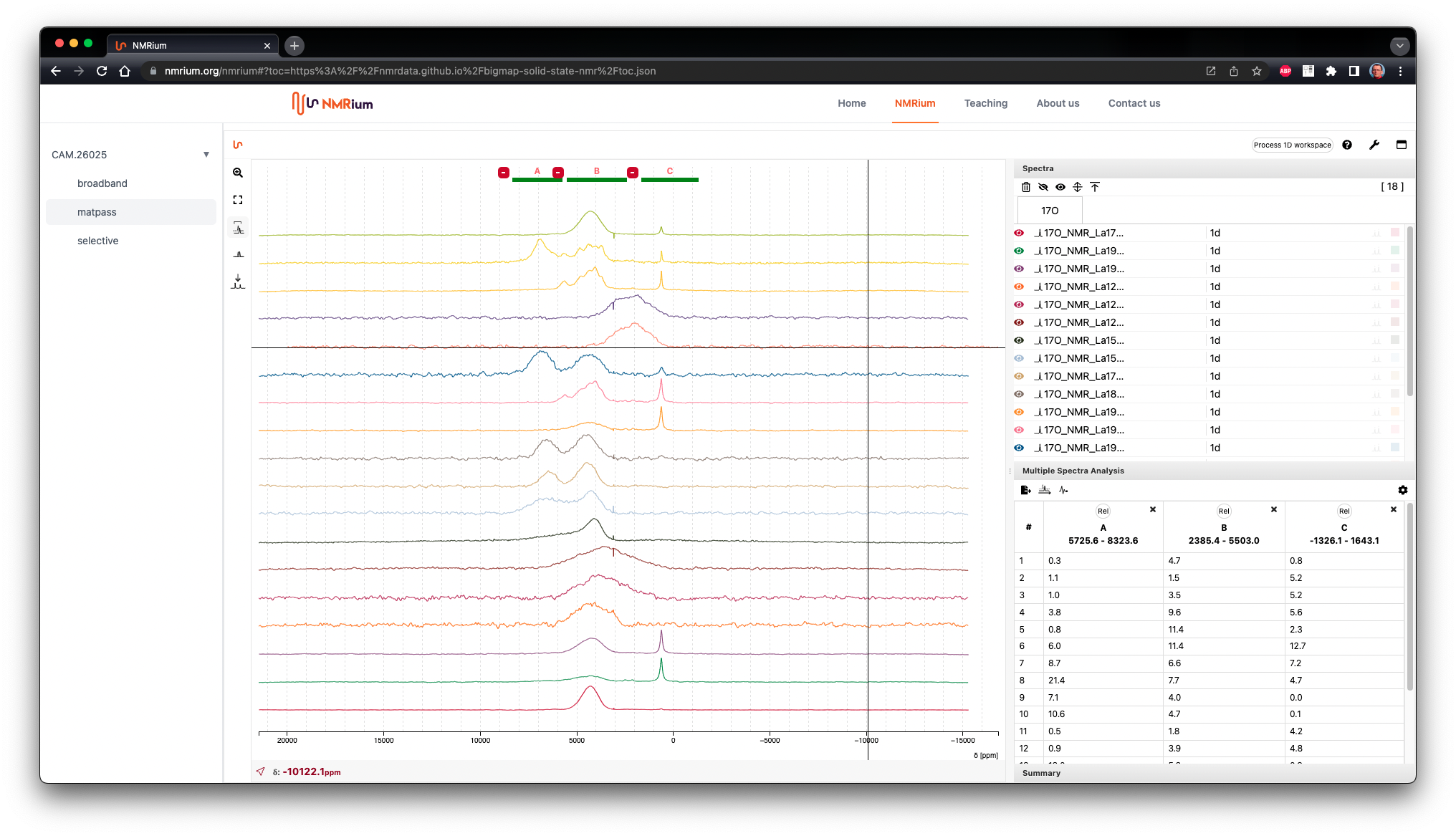
General overview of the features
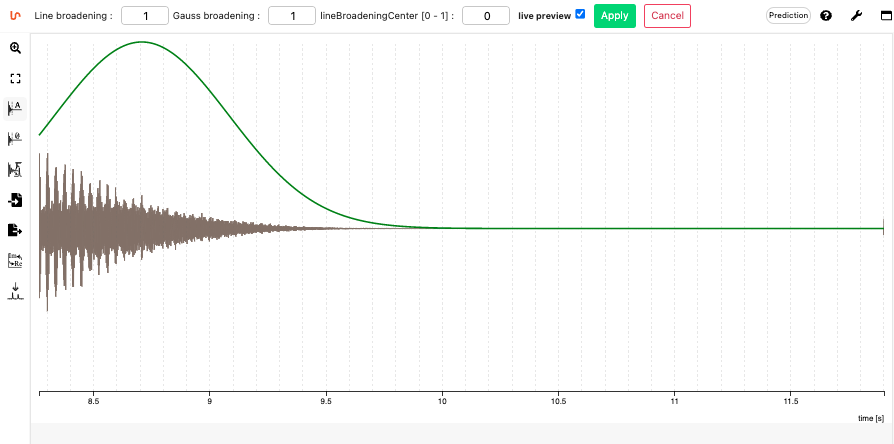
From the FID
NMRium allows to load native 1D data from Bruker (Zip file), Varian and Jeol as well as the JCAMP-DX file format. Either the FID or the FT file can be loaded.
From the FID the following operations can be done
- Apodization (Line broadening, Gauss broadening, Center for line broadening)
- Zero filling
- Fourier transform
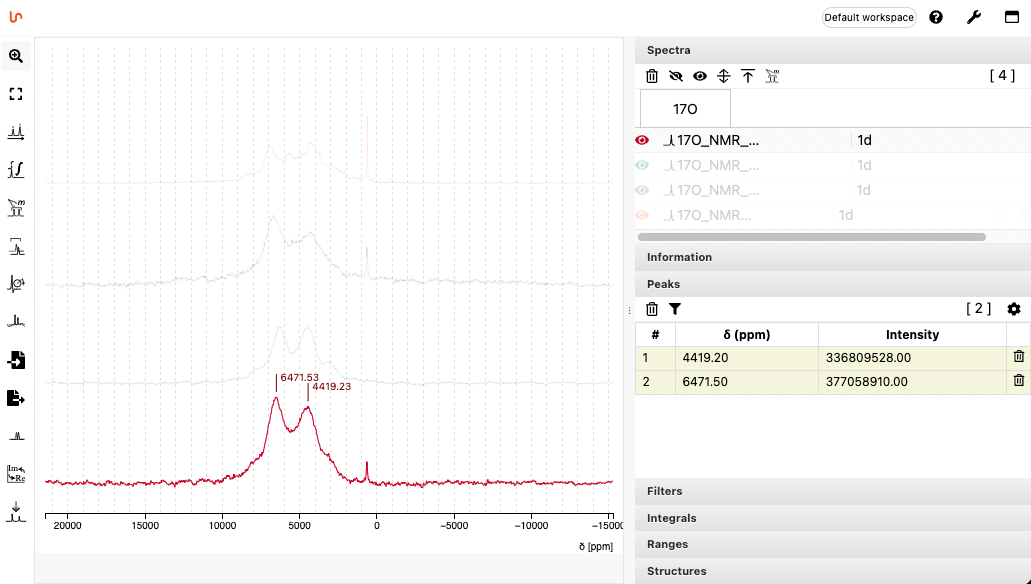
Analyze the spectrum
A tool allows to make peak picking and to reference the spectrum if required.
Exportation
The image of the spectrum can be copied using CTRLC but can also be downloaded to the native .nmrium format (for further processing) or as a svg to create high quality reports.
Integration if your own system
There are various ways to integrate NMRium in your own system, from a close integration using the React component to a simple link to www.nmrium.org.
Integration in your own React website
NMRium is published on npm and can be installed using:
npm i nmrium
It can then be included in your jsx component using the following syntax:
import NMRium from "nmrium";
function MyComponent() {
return <NMRium />;
}
More information about loading the data from a prop and using the numerous options can be found on https://docs.nmrium.org/for-developers/include-react-component.
Visualization of spectra on https://www.nmrium.org/nmrium
The website https://www.nmrium.org/nmrium provides a demonstration of NMRium component. On this page you are able either to drag and drop a list of JCAMP-DX or a zip file containing native Bruker files.
The use of NMRium is extensively described on https://docs.nmrium.org.
Opening directly your spectra from your ELN on https://www.nmrium.org
The website www.nmrium.org is able to load a Table Of Contents (TOC) as a JSON file.
If order to have a menu on the left containing many group of spectra you should create a correctly formatted .json file. In this case the URL to this JSON file has the following structure:
https://www.nmrium.org/nmrium#?json=jsonURL
The jsonURL must be accessible using Ajax (take care about cross-origin and https). This file could be generated dynamically directly from a database.
Here is an example of the content of the toc.json file:
[
{
"groupName": "CAM.26025",
"folderName": "CAM.26025",
"children": [
{
"id": "f3oJnR1zqfNNffI0Urjkf1oZifc=",
"file": "./CAM.26025/broadband/index.json",
"title": "broadband",
"selected": true
},
{
"id": "gcVdupbPgszMCpSM3lPoS/AcG3c=",
"file": "./CAM.26025/matpass/index.json",
"title": "matpass"
},
{
"id": "t87E7IvM5VKHAEjtX1Q2HCB23d4=",
"file": "./CAM.26025/selective/index.json",
"title": "selective"
}
]
}
]
The file attribute will contain the relative link to an NMRium file (encodes as a JSON) that describes the set of JCAMP-DX to load. Here is for example the content of the file ./CAM.26025/broadband/index.json:
{
"spectra": [
{
"source": {
"jcampURL": "./17O_NMR_La17Sr03NiO4_broadband-spinecho.dx"
},
"display": {
"name": "17O_NMR_La17Sr03NiO4_broadband-spinecho.dx"
}
},
{
"source": {
"jcampURL": "./17O_NMR_La18Sr02NiO4_broadband-spinecho.dx"
},
"display": {
"name": "17O_NMR_La18Sr02NiO4_broadband-spinecho.dx"
}
},
{
"source": {
"jcampURL": "./17O_NMR_La19Sr01NiO4_broadband-spinecho.dx"
},
"display": {
"name": "17O_NMR_La19Sr01NiO4_broadband-spinecho.dx"
}
},
{
"source": {
"jcampURL": "./17O_NMR_La2NiO4_broadband-spinecho.dx"
},
"display": {
"name": "17O_NMR_La2NiO4_broadband-spinecho.dx"
}
}
]
}
This example can be tested here: https://www.nmrium.org/nmrium#?toc=https%3A%2F%2Fcheminfo.github.io%2Fnmr-dataset3%2Fmultiplet%2Findex.json
More information can be found on https://docs.nmrium.org/for-developers/using-nmrium#loading-a-table-of-contents.
Creating your own TOC from command line
We have developed a utility that simplifies the process of creating a static set of spectra and the toc.json file.
The principle is quite simple, you create folders containing jcamp files (extension dx or jdx) and optionally a molfile (extension .mol).
If you have node installed, execute the following instructions to generate the correct json files:
npm i --global nmrium-cli
nmrium createGeneralTOC
This command will generate the required toc.json file as well as the corresponding .json in each folder. If this toc.json is accessible from a URL it can be loaded by NMRium as explained above.
Future developments
While NMRium fulfill the requirements to visualize and process solid state NMR spectra, more tools can always be implemented and could be part of another project.
In particular we would like to focus on:
- peak shape analysis
- multi spectra analysis and difference in spectra
- reference database for quick and easy comparison of other spectra
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 957189.
